What Is The Main Function Of The Lysosomes In An Animal Cell?
| Jail cell biology | |
|---|---|
| Animal prison cell diagram | |
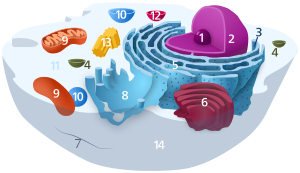 Components of a typical brute cell:
|
A lysosome () is a membrane-bound organelle found in many animal cells.[1] They are spherical vesicles that contain hydrolytic enzymes that can intermission down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane proteins, and its lumenal proteins. The lumen's pH (~4.five–5.0)[2] is optimal for the enzymes involved in hydrolysis, coordinating to the activity of the stomach. Besides degradation of polymers, the lysosome is involved in various jail cell processes, including secretion, plasma membrane repair, apoptosis, jail cell signaling, and energy metabolism.[3]

Lysosomes digest materials taken into the cell and recycle intracellular materials. Stride 1 shows fabric entering a food vacuole through the plasma membrane, a process known every bit endocytosis. In step two a lysosome with an agile hydrolytic enzyme comes into the picture as the food vacuole moves abroad from the plasma membrane. Step iii consists of the lysosome fusing with the food vacuole and hydrolytic enzymes inbound the food vacuole. In the final stride, stride four, hydrolytic enzymes assimilate the food particles.[four]
Lysosomes act as the waste disposal system of the cell past digesting used materials in the cytoplasm, from both inside and outside the jail cell. Material from outside the cell is taken up through endocytosis, while textile from the inside of the cell is digested through autophagy.[5] The sizes of the organelles vary greatly—the larger ones can exist more than 10 times the size of the smaller ones.[6] They were discovered and named by Belgian biologist Christian de Duve, who somewhen received the Nobel Prize in Physiology or Medicine in 1974.
Lysosomes are known to contain more than lx unlike enzymes, and take more than l membrane proteins.[seven] [eight] Enzymes of the lysosomes are synthesised in the rough endoplasmic reticulum and exported to the Golgi apparatus upon recruitment by a circuitous composed of CLN6 and CLN8 proteins.[nine] [10] The enzymes are trafficked from the Golgi appliance to lysosomes in small vesicles, which fuse with larger acidic vesicles. Enzymes destined for a lysosome are specifically tagged with the molecule mannose half dozen-phosphate, and then that they are properly sorted into acidified vesicles.[xi] [12]
In 2009, Marco Sardiello and co-workers discovered that the synthesis of most lysosomal enzymes and membrane proteins is controlled by transcription factor EB (TFEB), which promotes the transcription of nuclear genes.[13] [fourteen] Mutations in the genes for these enzymes are responsible for more than l unlike human genetic disorders, which are collectively known every bit lysosomal storage diseases. These diseases result from an accumulation of specific substrates, due to the inability to break them downward. These genetic defects are related to several neurodegenerative disorders, cancers, cardiovascular diseases, and crumbling-related diseases.[15] [16] [17]
Discovery [edit]

TEM views of diverse vesicular compartments. Lysosomes are denoted past "Ly". They are dyed dark due to their acidity; in the center of the top paradigm, a Golgi Apparatus tin can be seen, distal from the cell membrane relative to the lysosomes.
Christian de Duve, the chairman of the Laboratory of Physiological Chemistry at the Catholic University of Louvain in Belgium, had been studying the machinery of action of a pancreatic hormone insulin in liver cells. By 1949, he and his team had focused on the enzyme chosen glucose 6-phosphatase, which is the first crucial enzyme in sugar metabolism and the target of insulin. They already suspected that this enzyme played a key role in regulating claret sugar levels. However, fifty-fifty later on a serial of experiments, they failed to purify and isolate the enzyme from the cellular extracts. Therefore, they tried a more backbreaking procedure of cell fractionation, by which cellular components are separated based on their sizes using centrifugation.
They succeeded in detecting the enzyme activity from the microsomal fraction. This was the crucial step in the serendipitous discovery of lysosomes. To gauge this enzyme activity, they used that of the standardized enzyme acrid phosphatase and found that the activity was just x% of the expected value. One day, the enzyme activity of purified jail cell fractions which had been refrigerated for five days was measured. Surprisingly, the enzyme activity was increased to normal of that of the fresh sample. The result was the same no matter how many times they repeated the estimation, and led to the decision that a membrane-like bulwark limited the accessibility of the enzyme to its substrate, and that the enzymes were able to diffuse later on a few days (and react with their substrate). They described this membrane-similar bulwark equally a "saclike structure surrounded by a membrane and containing acrid phosphatase."[xviii]
It became articulate that this enzyme from the prison cell fraction came from bleary fractions, which were definitely prison cell organelles, and in 1955 De Duve named them "lysosomes" to reverberate their digestive properties.[19] The same year, Alex B. Novikoff from the University of Vermont visited de Duve's laboratory, and successfully obtained the get-go electron micrographs of the new organelle. Using a staining method for acid phosphatase, de Duve and Novikoff confirmed the location of the hydrolytic enzymes of lysosomes using light and electron microscopic studies.[20] [21] de Duve won the Nobel Prize in Physiology or Medicine in 1974 for this discovery.
Originally, De Duve had termed the organelles the "suicide bags" or "suicide sacs" of the cells, for their hypothesized function in apoptosis.[22] However, it has since been concluded that they just play a minor office in cell expiry.[23]
Function and structure [edit]
Lysosomes comprise a diverseness of enzymes, enabling the cell to break down various biomolecules it engulfs, including peptides, nucleic acids, carbohydrates, and lipids (lysosomal lipase). The enzymes responsible for this hydrolysis require an acidic environment for optimal action.
In addition to being able to break downwards polymers, lysosomes are capable of fusing with other organelles & digesting large structures or cellular debris; through cooperation with phagosomes, they are able to conduct autophagy, clearing out damaged structures. Similarly, they are able to intermission down virus particles or bacteria in phagocytosis of macrophages.
The size of lysosomes varies from 0.1 μm to 1.2 μm.[24] With a pH ranging from ~4.5–5.0, the interior of the lysosomes is acidic compared to the slightly bones cytosol (pH 7.2). The lysosomal membrane protects the cytosol, and therefore the rest of the cell, from the degradative enzymes within the lysosome. The cell is additionally protected from any lysosomal acid hydrolases that drain into the cytosol, equally these enzymes are pH-sensitive and practise not function well or at all in the element of group i environment of the cytosol. This ensures that cytosolic molecules and organelles are not destroyed in example at that place is leakage of the hydrolytic enzymes from the lysosome.
The lysosome maintains its pH differential past pumping in protons (H+ ions) from the cytosol across the membrane via proton pumps and chloride ion channels. Vacuolar-ATPases are responsible for transport of protons, while the counter ship of chloride ions is performed by ClC-7 Cl−/H+ antiporter. In this way a steady acidic environment is maintained.[25] [26]
Information technology sources its versatile capacity for degradation by import of enzymes with specificity for unlike substrates; cathepsins are the major class of hydrolytic enzymes, while lysosomal blastoff-glucosidase is responsible for carbohydrates, and lysosomal acrid phosphatase is necessary to release phosphate groups of phospholipids.
Germination [edit]

The lysosome is shown in purple, equally an endpoint in endocytotic sorting. AP2 is necessary for vesicle germination, whereas the mannose-half dozen-receptor is necessary for sorting hydrolase into the lysosome'due south lumen.
Many components of brute cells are recycled by transferring them inside or embedded in sections of membrane. For example, in endocytosis (more specifically, macropinocytosis), a portion of the cell's plasma membrane pinches off to form vesicles that will somewhen fuse with an organelle inside the cell. Without active replenishment, the plasma membrane would continuously decrease in size. It is idea that lysosomes participate in this dynamic membrane substitution system and are formed past a gradual maturation process from endosomes.[27] [28]
The production of lysosomal proteins suggests one method of lysosome sustainment. Lysosomal protein genes are transcribed in the nucleus in a process that is controlled by transcription gene EB (TFEB).[14] mRNA transcripts exit the nucleus into the cytosol, where they are translated by ribosomes. The nascent peptide chains are translocated into the rough endoplasmic reticulum, where they are modified. Lysosomal soluble proteins get out the endoplasmic reticulum via COPII-coated vesicles afterwards recruitment past the EGRESS circuitous (ER-to-Grandolgi relaying of enzymes of the lysosomal system), which is composed of CLN6 and CLN8 proteins.[9] [10] COPII vesicles then deliver lysosomal enzymes to the Golgi appliance, where a specific lysosomal tag, mannose 6-phosphate, is added to the peptides. The presence of these tags allow for binding to mannose 6-phosphate receptors in the Golgi apparatus, a phenomenon that is crucial for proper packaging into vesicles destined for the lysosomal system.[29]
Upon leaving the Golgi apparatus, the lysosomal enzyme-filled vesicle fuses with a belatedly endosome, a relatively acidic organelle with an approximate pH of 5.5. This acidic environment causes dissociation of the lysosomal enzymes from the mannose vi-phosphate receptors. The enzymes are packed into vesicles for farther ship to established lysosomes.[29] The late endosome itself can somewhen grow into a mature lysosome, equally evidenced past the transport of endosomal membrane components from the lysosomes back to the endosomes.[27]
Pathogen entry [edit]

Cholera gaining entry into a prison cell via endocytosis.
As the endpoint of endocytosis, the lysosome also acts as a safeguard in preventing pathogens from beingness able to accomplish the cytoplasm before being degraded. Pathogens oft hijack endocytotic pathways such as pinocytosis in order to proceeds entry into the cell. The lysosome prevents piece of cake entry into the prison cell by hydrolyzing the biomolecules of pathogens necessary for their replication strategies; reduced Lysosomal action results in an increase in viral infectivity, including HIV.[30] In addition, ABfive toxins such as cholera hijack the endosomal pathway while evading lysosomal degradation.[xxx]
Clinical significance [edit]
Lysosomes are involved in a group of genetically inherited deficiencies, or mutations chosen lysosomal storage diseases (LSD), inborn errors of metabolism caused by a dysfunction of one of the enzymes. The rate of incidence is estimated to exist 1 in 5,000 births, and the truthful effigy expected to be college every bit many cases are likely to be undiagnosed or misdiagnosed. The primary cause is deficiency of an acid hydrolase. Other conditions are due to defects in lysosomal membrane proteins that fail to transport the enzyme, non-enzymatic soluble lysosomal proteins. The initial effect of such disorders is aggregating of specific macromolecules or monomeric compounds inside the endosomal–autophagic–lysosomal arrangement.[xv] This results in aberrant signaling pathways, calcium homeostasis, lipid biosynthesis and degradation and intracellular trafficking, ultimately leading to pathogenetic disorders. The organs near affected are brain, viscera, bone and cartilage.[31] [32]
At that place is no direct medical treatment to cure LSDs.[33] The most common LSD is Gaucher's disease, which is due to deficiency of the enzyme glucocerebrosidase. Consequently, the enzyme substrate, the fatty acid glucosylceramide accumulates, particularly in white blood cells, which in turn affects spleen, liver, kidneys, lungs, brain and os marrow. The disease is characterized by bruises, fatigue, anaemia, low claret platelets, osteoporosis, and enlargement of the liver and spleen.[34] [35] As of 2017, enzyme replacement therapy is bachelor for treating eight of the 50-60 known LDs.[36]
The most severe and rarely found, lysosomal storage disease is inclusion jail cell disease.[37]
Metachromatic leukodystrophy is another lysosomal storage disease that also affects sphingolipid metabolism.
Dysfunctional lysosome activeness is likewise heavily implicated in the biological science of crumbling, and age-related diseases such as Alzheimer'southward, Parkinson's, and cardiovascular illness. [38] [39]
Different enzymes present in Lysosomes [xl] [edit]
| Sr. No | Enzymes | Substrate |
|---|---|---|
| 1 | Phosphates | |
| A- Acid phosphatase | Most phosphomonoesters | |
| B- Acid phosphodiesterase | Oligonucleotides and phosphodiesterase | |
| 2 | Nucleases | |
| A- Acid ribonuclease | RNA | |
| B- Acid deoxyribonuclease | DNA | |
| 3 | Polysaccharides/ mucopolysaccharides hydrolyzing enzymes | |
| A- β-Galactosidase | Galactosides | |
| B- α-Glucosidase | Glycogen | |
| C- α-Mannosidase | Mannosides, glycoproteins | |
| D- β- Glucoronidase | Polysaccharides and mucopolysaccharides | |
| Eastward- Lysozymes | Bacterial jail cell walls and mucopolysaccharides | |
| F- Hyaluronidase | Hyaluronic acids, chondroitin sulfates | |
| H- Arylsulphatase | Organic sulfates | |
| 4 | Proteases | |
| A- Cathepsin(south) | Proteins | |
| B- Collagenase | Collagen | |
| C- Peptidase | Peptides | |
| 5 | Lipid degrading enzymes | |
| A- Esterase | Fat acyl esters | |
| B- Phospholipase | Phospholipids | |
| 6 | Sulfatases | |
| A- Arylsulfatase(A, B & G) | O- and N-Sulfate esters | |
| B- Glucosamine (N-acetyl)-six-Sulfatase/GNS | Glycosaminoglycans | |
| C- Iduronate 2-Sulfatase/IDS | O- and N-Sulfate esters |
Lysosomotropism [edit]
Weak bases with lipophilic properties accumulate in acidic intracellular compartments like lysosomes. While the plasma and lysosomal membranes are permeable for neutral and uncharged species of weak bases, the charged protonated species of weak bases do not permeate biomembranes and accrue within lysosomes. The concentration inside lysosomes may accomplish levels 100 to g fold college than extracellular concentrations. This phenomenon is chosen lysosomotropism,[41] "acid trapping" or "proton pump" upshot.[42] The amount of accumulation of lysosomotropic compounds may be estimated using a cell-based mathematical model.[43]
A significant role of the clinically canonical drugs are lipophilic weak bases with lysosomotropic properties. This explains a number of pharmacological properties of these drugs, such every bit high tissue-to-blood concentration gradients or long tissue elimination one-half-lives; these properties have been found for drugs such as haloperidol,[44] levomepromazine,[45] and amantadine.[46] Yet, high tissue concentrations and long emptying half-lives are explained also by lipophilicity and absorption of drugs to fatty tissue structures. Of import lysosomal enzymes, such as acid sphingomyelinase, may exist inhibited by lysosomally accumulated drugs.[47] [48] Such compounds are termed FIASMAs (functional inhibitor of acid sphingomyelinase)[49] and include for instance fluoxetine, sertraline, or amitriptyline.
Ambroxol is a lysosomotropic drug of clinical use to care for atmospheric condition of productive cough for its mucolytic activity. Ambroxol triggers the exocytosis of lysosomes via neutralization of lysosomal pH and calcium release from acidic calcium stores.[fifty] Presumably for this reason, Ambroxol was also found to meliorate cellular part in some disease of lysosomal origin such as Parkinson's or lysosomal storage disease.[51] [52]
Systemic lupus erythematosus [edit]
Impaired lysosome function is prominent in systemic lupus erythematosus preventing macrophages and monocytes from degrading neutrophil extracellular traps[53] and immune complexes.[54] [55] [56] The failure to degrade internalized immune complexes stems from chronic mTORC2 activity, which impairs lysosome acidification.[57] As a consequence, allowed complexes in the lysosome recycle to the surface of macrophages causing an accumulation of nuclear antigens upstream of multiple lupus-associated pathologies.[54] [58] [59]
Controversy in botany [edit]
Past scientific convention, the term lysosome is applied to these vesicular organelles just in animals, and the term vacuole is applied to those in plants, fungi and algae (some animate being cells also have vacuoles). Discoveries in plant cells since the 1970s started to challenge this definition. Plant vacuoles are institute to be much more diverse in structure and office than previously idea.[lx] [61] Some vacuoles contain their own hydrolytic enzymes and perform the archetype lysosomal activity, which is autophagy.[62] [63] [64] These vacuoles are therefore seen as fulfilling the office of the animal lysosome. Based on de Duve's clarification that "only when considered every bit part of a arrangement involved directly or indirectly in intracellular digestion does the term lysosome describe a physiological unit", some botanists strongly argued that these vacuoles are lysosomes.[65] However, this is not universally accepted as the vacuoles are strictly not similar to lysosomes, such equally in their specific enzymes and lack of phagocytic functions.[66] Vacuoles do not take catabolic activity and do not undergo exocytosis as lysosomes do.[67]
Etymology and pronunciation [edit]
The give-and-take lysosome (, ) is New Latin that uses the combining forms lyso- (referring to lysis and derived from the Latin lysis, significant "to loosen", via Ancient Greek λύσις [lúsis]), and -some, from soma, "trunk", yielding "body that lyses" or "lytic trunk". The adjectival form is lysosomal. The forms *lyosome and *lyosomal are much rarer; they use the lyo- form of the prefix but are ofttimes treated by readers and editors as mere unthinking replications of typos, which has no dubiousness been truthful as often every bit not.
Run across also [edit]
- Peroxisome
- Cathelicidin
- Antimicrobial peptides
- Innate allowed organization
References [edit]
- ^ By convention like cells in plants are chosen vacuoles, encounter § Controversy in phytology
- ^ Ohkuma Due south, Poole B (July 1978). "Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents". Proceedings of the National Academy of Sciences of the The states of America. 75 (vii): 3327–31. Bibcode:1978PNAS...75.3327O. doi:ten.1073/pnas.75.7.3327. PMC392768. PMID 28524.
- ^ Settembre C, Fraldi A, Medina DL, Ballabio A (May 2013). "Signals from the lysosome: a control centre for cellular clearance and energy metabolism". Nature Reviews Molecular Cell Biology. 14 (5): 283–96. doi:10.1038/nrm3565. PMC4387238. PMID 23609508.
- ^ Holtzclaw FW, et al. (2008). AP* Biology: to Back-trail Biology (8th AP ed.). Pearson Benjamin Cummings.
- ^ Underwood, Emily (2018). "When the brain's waste disposal system fails". Knowable Magazine. doi:x.1146/knowable-121118-one.
- ^ Lüllmznn-Rauch R (2005). "History and Morphology of Lysosome". In Zaftig P (ed.). Lysosomes (Online-Ausg. 1 ed.). Georgetown, Tex.: Landes Bioscience/Eurekah.com. pp. 1–sixteen. ISBN978-0-387-28957-ane.
- ^ Xu H, Ren D (2015). "Lysosomal physiology". Annual Review of Physiology. 77 (1): 57–80. doi:10.1146/annurev-physiol-021014-071649. PMC4524569. PMID 25668017.
- ^ "Lysosomal Enzymes". www.rndsystems.com. R&D Systems. Retrieved iv Oct 2016.
- ^ a b di Ronza A, Bajaj Fifty, Sharma J, Sanagasetti D, Lotfi P, Adamski CJ, Collette J, Palmieri M, Amawi A, Popp L, Chang KT, Meschini MC, Leung HE, Segatori L, Simonati A, Sifers RN, Santorelli FM, Sardiello M (December 2018). "CLN8 is an endoplasmic reticulum cargo receptor that regulates lysosome biogenesis". Nature Cell Biology. twenty (12): 1370–1377. doi:ten.1038/s41556-018-0228-7. PMC6277210. PMID 30397314.
- ^ a b Bajaj L, Sharma J, di Ronza A, Zhang P, Eblimit A, Pal R, Roman D, Collette JR, Booth C, Chang KT, Sifers RN, Jung SY, Weimer JM, Chen R, Schekman RW, Sardiello M (June 2020). "A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer". J Clin Invest. 130 (8): 4118–4132. doi:10.1172/JCI130955. PMC7410054. PMID 32597833.
- ^ Saftig P, Klumperman J (September 2009). "Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function". Nature Reviews Molecular Cell Biology. x (ix): 623–35. doi:10.1038/nrm2745. PMID 19672277. S2CID 24493663.
- ^ Samie MA, Xu H (June 2014). "Lysosomal exocytosis and lipid storage disorders". Journal of Lipid Research. 55 (6): 995–1009. doi:10.1194/jlr.R046896. PMC4031951. PMID 24668941.
- ^ Underwood, Emily (2018). "When the encephalon's waste disposal system fails". Knowable Magazine. doi:10.1146/knowable-121118-1.
- ^ a b Sardiello 1000, Palmieri Chiliad, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione 5, Polishchuk RS, Banfi S, Parenti 1000, Cattaneo E, Ballabio A (July 2009). "A cistron network regulating lysosomal biogenesis and function". Scientific discipline. 325 (5939): 473–7. Bibcode:2009Sci...325..473S. doi:10.1126/scientific discipline.1174447. PMID 19556463. S2CID 20353685.
- ^ a b Platt FM, Boland B, van der Spoel Ac (Nov 2012). "The jail cell biology of disease: lysosomal storage disorders: the cellular touch of lysosomal dysfunction". The Journal of Prison cell Biology. 199 (5): 723–34. doi:10.1083/jcb.201208152. PMC3514785. PMID 23185029.
- ^ He LQ, Lu JH, Yue ZY (May 2013). "Autophagy in ageing and ageing-associated diseases". Acta Pharmacologica Sinica. 34 (5): 605–11. doi:10.1038/aps.2012.188. PMC3647216. PMID 23416930.
- ^ Carmona-Gutierrez, Didac; Hughes, Adam L.; Madeo, Frank; Ruckenstuhl, Christoph (1 December 2016). "The crucial affect of lysosomes in crumbling and longevity". Ageing Enquiry Reviews. Lysosomes in Aging. 32: 2–12. doi:ten.1016/j.arr.2016.04.009. ISSN 1568-1637. PMC5081277. PMID 27125853.
- ^ Susana Castro-Obregon (2010). "The Discovery of Lysosomes and Autophagy". Nature Education. iii (9): 49.
- ^ de Duve C (September 2005). "The lysosome turns fifty". Nature Jail cell Biology. 7 (9): 847–9. doi:10.1038/ncb0905-847. PMID 16136179. S2CID 30307451.
- ^ Novikoff AB, Beaufay H, De Duve C (July 1956). "Electron microscopy of lysosomerich fractions from rat liver". The Journal of Biophysical and Biochemical Cytology. ii (iv Suppl): 179–84. doi:x.1083/jcb.2.four.179. PMC2229688. PMID 13357540.
- ^ Klionsky DJ (August 2008). "Autophagy revisited: a chat with Christian de Duve". Autophagy. 4 (six): 740–three. doi:ten.4161/auto.6398. PMID 18567941.
- ^ Hayashi, Teru, and others. "Subcellular Particles." Subcellular Particles., 1959.
- ^ Turk B, Turk V (2009). "Lysosomes as 'Suicide Numberless' in Cell Death: Myth or Reality?". The Journal of Biological Chemical science. 284 (33): 21783–87. doi:ten.1074/jbc.R109.023820. PMC2755904. PMID 19473965.
- ^ Kuehnel W (2003). Color Atlas of Cytology, Histology, & Microscopic Anatomy (fourth ed.). Thieme. p. 34. ISBN978-1-58890-175-0.
- ^ Mindell JA (2012). "Lysosomal acidification mechanisms". Annual Review of Physiology. 74 (1): 69–86. doi:10.1146/annurev-physiol-012110-142317. PMID 22335796.
- ^ Ishida Y, Nayak S, Mindell JA, Grabe Thou (June 2013). "A model of lysosomal pH regulation". The Periodical of General Physiology. 141 (6): 705–xx. doi:10.1085/jgp.201210930. PMC3664703. PMID 23712550.
- ^ a b Alberts B, et al. (2002). Molecular biological science of the prison cell (4th ed.). New York: Garland Science. ISBN978-0-8153-3218-3.
- ^ Falcone South, Cocucci E, Podini P, Kirchhausen T, Clementi Due east, Meldolesi J (November 2006). "Macropinocytosis: regulated coordination of endocytic and exocytic membrane traffic events". Journal of Jail cell Science. 119 (Pt 22): 4758–69. doi:x.1242/jcs.03238. PMID 17077125.
- ^ a b Lodish H, et al. (2000). Molecular cell biological science (fourth ed.). New York: Scientific American Books. ISBN978-0-7167-3136-8.
- ^ a b Wei BL, Denton PW, O'Neill E, Luo T, Foster JL, Garcia JV (May 2005). "Inhibition of lysosome and proteasome function enhances human being immunodeficiency virus blazon 1 infection". Journal of Virology. 79 (9): 5705–12. doi:10.1128/jvi.79.9.5705-5712.2005. PMC1082736. PMID 15827185.
- ^ Schultz ML, Tecedor L, Chang One thousand, Davidson BL (Baronial 2011). "Clarifying lysosomal storage diseases". Trends in Neurosciences. 34 (8): 401–10. doi:ten.1016/j.tins.2011.05.006. PMC3153126. PMID 21723623.
- ^ Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A (May 2012). "Autophagy in lysosomal storage disorders". Autophagy. viii (five): 719–thirty. doi:10.4161/auto.19469. PMC3378416. PMID 22647656.
- ^ .Parenti G, Pignata C, Vajro P, Salerno M (January 2013). "New strategies for the treatment of lysosomal storage diseases (review)". International Journal of Molecular Medicine. 31 (1): xi–xx. doi:10.3892/ijmm.2012.1187. PMID 23165354.
- ^ Rosenbloom Be, Weinreb NJ (2013). "Gaucher illness: a comprehensive review". Critical Reviews in Oncogenesis. xviii (iii): 163–75. doi:10.1615/CritRevOncog.2013006060. PMID 23510062.
- ^ Sidransky E (October 2012). "Gaucher affliction: insights from a rare Mendelian disorder". Discovery Medicine. 14 (77): 273–81. PMC4141347. PMID 23114583.
- ^ Solomon M, Muro S (September 2017). "Lysosomal enzyme replacement therapies: Historical development, clinical outcomes, and future perspectives". Avant-garde Drug Delivery Reviews. 118: 109–134. doi:10.1016/j.addr.2017.05.004. PMC5828774. PMID 28502768.
- ^ Alberts, Bruce (2002). Molecular biology of the prison cell (4th ed.). Garland Scientific discipline. p. 744. ISBN978-0815340720.
- ^ Carmona-Gutierrez, Didac; Hughes, Adam L.; Madeo, Frank; Ruckenstuhl, Christoph (ane December 2016). "The crucial impact of lysosomes in crumbling and longevity". Ageing Enquiry Reviews. Lysosomes in Crumbling. 32: ii–12. doi:ten.1016/j.arr.2016.04.009. ISSN 1568-1637. PMC5081277. PMID 27125853.
- ^ Finkbeiner, Steven (1 Apr 2019). "The Autophagy Lysosomal Pathway and Neurodegeneration". Cold Spring Harbor Perspectives in Biology. 12 (three): a033993. doi:ten.1101/cshperspect.a033993. ISSN 1943-0264. PMC6773515. PMID 30936119.
- ^ Pranav Kumar. (2013). Life Sciences : Fundamentals and practice. Mina, Usha. (3rd ed.). New Delhi: Pathfinder Academy. ISBN978-81-906427-0-v. OCLC 857764171.
- ^ de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F (September 1974). "Commentary. Lysosomotropic agents". Biochemical Pharmacology. 23 (18): 2495–531. doi:ten.1016/0006-2952(74)90174-nine. PMID 4606365.
- ^ Traganos F, Darzynkiewicz Z (1994). "Lysosomal proton pump action: supravital cell staining with acridine orangish differentiates leukocyte subpopulations". Methods Cell Biol. 41: 185–94. doi:10.1016/S0091-679X(08)61717-3. PMID 7532261.
- ^ Trapp South, Rosania GR, Horobin RW, Kornhuber J (October 2008). "Quantitative modeling of selective lysosomal targeting for drug pattern". European Biophysics Periodical. 37 (viii): 1317–28. doi:10.1007/s00249-008-0338-4. PMC2711917. PMID 18504571.
- ^ Kornhuber J, Schultz A, Wiltfang J, Meineke I, Gleiter CH, Zöchling R, Boissl KW, Leblhuber F, Riederer P (June 1999). "Persistence of haloperidol in homo encephalon tissue". The American Periodical of Psychiatry. 156 (six): 885–90. doi:10.1176/ajp.156.6.885. PMID 10360127. S2CID 7258546.
- ^ Kornhuber J, Weigmann H, Röhrich J, Wiltfang J, Bleich S, Meineke I, Zöchling R, Härtter S, Riederer P, Hiemke C (March 2006). "Region specific distribution of levomepromazine in the human being brain". Journal of Neural Transmission. 113 (3): 387–97. doi:10.1007/s00702-005-0331-3. PMID 15997416. S2CID 24735371.
- ^ Kornhuber J, Quack G, Danysz Due west, Jellinger Yard, Danielczyk Westward, Gsell Due west, Riederer P (July 1995). "Therapeutic brain concentration of the NMDA receptor adversary amantadine". Neuropharmacology. 34 (7): 713–21. doi:10.1016/0028-3908(95)00056-c. PMID 8532138. S2CID 25784783.
- ^ Kornhuber J, Tripal P, Reichel M, Terfloth L, Bleich Southward, Wiltfang J, Gulbins E (January 2008). "Identification of new functional inhibitors of acid sphingomyelinase using a construction-property-activity relation model". Journal of Medicinal Chemical science. 51 (ii): 219–37. CiteSeerX10.1.ane.324.8854. doi:10.1021/jm070524a. PMID 18027916.
- ^ Kornhuber J, Muehlbacher M, Trapp South, Pechmann Southward, Friedl A, Reichel M, Mühle C, Terfloth 50, Groemer TW, Spitzer GM, Liedl KR, Gulbins E, Tripal P (2011). Riezman H (ed.). "Identification of novel functional inhibitors of acrid sphingomyelinase". PLOS Ane. six (8): e23852. Bibcode:2011PLoSO...623852K. doi:ten.1371/periodical.pone.0023852. PMC3166082. PMID 21909365.
- ^ Kornhuber J, Tripal P, Reichel M, Mühle C, Rhein C, Muehlbacher M, Groemer TW, Gulbins E (2010). "Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): a novel pharmacological grouping of drugs with broad clinical applications". Cellular Physiology and Biochemistry. 26 (1): ix–20. doi:x.1159/000315101. PMID 20502000.
- ^ Fois G, Hobi N, Felder E, Ziegler A, Miklavc P, Walther P, Radermacher P, Haller T, Dietl P (Dec 2015). "A new part for an erstwhile drug: Ambroxol triggers lysosomal exocytosis via pH-dependent Ca²⁺ release from acidic Ca²⁺ stores". Cell Calcium. 58 (6): 628–37. doi:x.1016/j.ceca.2015.10.002. PMID 26560688.
- ^ Albin RL, Dauer WT (May 2014). "Magic shotgun for Parkinson's disease?". Encephalon. 137 (Pt five): 1274–5. doi:ten.1093/encephalon/awu076. PMID 24771397.
- ^ McNeill A, Magalhaes J, Shen C, Chau KY, Hughes D, Mehta A, Foltynie T, Cooper JM, Abramov AY, Gegg M, Schapira AH (May 2014). "Ambroxol improves lysosomal biochemistry in glucocerebrosidase mutation-linked Parkinson disease cells". Brain. 137 (Pt 5): 1481–95. doi:10.1093/encephalon/awu020. PMC3999713. PMID 24574503.
- ^ Hakkim A, Fürnrohr BG, Amann K, Laube B, Abed UA, Brinkmann Five, Herrmann 1000, Voll RE, Zychlinsky A (May 2010). "Damage of neutrophil extracellular trap degradation is associated with lupus nephritis". Proceedings of the National Academy of Sciences of the Us. 107 (21): 9813–viii. Bibcode:2010PNAS..107.9813H. doi:x.1073/pnas.0909927107. PMC2906830. PMID 20439745.
- ^ a b Monteith AJ, Kang S, Scott E, Hillman One thousand, Rajfur Z, Jacobson K, Costello MJ, Vilen BJ (April 2016). "Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus". Proceedings of the National Academy of Sciences of the United States of America. 113 (15): E2142–51. Bibcode:2016PNAS..113E2142M. doi:10.1073/pnas.1513943113. PMC4839468. PMID 27035940.
- ^ Kavai G, Szegedi G (August 2007). "Immune complex clearance by monocytes and macrophages in systemic lupus erythematosus". Autoimmunity Reviews. half-dozen (seven): 497–502. doi:10.1016/j.autrev.2007.01.017. PMID 17643939.
- ^ Kávai M, Csipö I, Sonkoly I, Csongor J, Szegedi GY (November 1986). "Defective immune circuitous degradation by monocytes in patients with systemic lupus erythematosus". Scandinavian Journal of Immunology. 24 (five): 527–32. doi:10.1111/j.1365-3083.1986.tb02167.x. PMID 3787186. S2CID 23685272.
- ^ Monteith AJ, Vincent HA, Kang Southward, Li P, Claiborne TM, Rajfur Z, Jacobson K, Moorman NJ, Vilen BJ (July 2018). "mTORC2 Action Disrupts Lysosome Acidification in Systemic Lupus Erythematosus by Impairing Caspase-1 Cleavage of Rab39a". Journal of Immunology. 201 (2): 371–382. doi:10.4049/jimmunol.1701712. PMC6039264. PMID 29866702.
- ^ Kang S, Rogers JL, Monteith AJ, Jiang C, Schmitz J, Clarke SH, Tarrant TK, Truong YK, Diaz G, Fedoriw Y, Vilen BJ (May 2016). "Apoptotic Debris Accumulates on Hematopoietic Cells and Promotes Affliction in Murine and Human Systemic Lupus Erythematosus". Journal of Immunology. 196 (10): 4030–9. doi:10.4049/jimmunol.1500418. PMC4868781. PMID 27059595.
- ^ Kang Due south, Fedoriw Y, Brenneman EK, Truong YK, Kikly K, Vilen BJ (April 2017). "BAFF Induces 3rd Lymphoid Structures and Positions T Cells within the Glomeruli during Lupus Nephritis". Periodical of Immunology. 198 (7): 2602–2611. doi:10.4049/jimmunol.1600281. PMC5360485. PMID 28235864.
- ^ Marty F (April 1999). "Plant vacuoles". The Institute Cell. 11 (4): 587–600. doi:10.2307/3870886. JSTOR 3870886. PMC144210. PMID 10213780.
- ^ Samaj J, Read ND, Volkmann D, Menzel D, Baluska F (August 2005). "The endocytic network in plants". Trends in Cell Biology. xv (8): 425–33. doi:10.1016/j.tcb.2005.06.006. PMID 16006126.
- ^ Matile, P (1978). "Biochemistry and Function of Vacuoles". Annual Review of Constitute Physiology. 29 (1): 193–213. doi:10.1146/annurev.pp.29.060178.001205.
- ^ Moriyasu Y, Ohsumi Y (August 1996). "Autophagy in Tobacco Suspension-Cultured Cells in Response to Sucrose Starvation". Plant Physiology. 111 (four): 1233–1241. doi:10.1104/pp.111.4.1233. PMC161001. PMID 12226358.
- ^ Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH (January 2012). "A novel poly peptide RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice". Molecular Establish. v (i): 205–17. doi:10.1093/mp/ssr081. PMID 21980143.
- ^ Swanson SJ, Bethke PC, Jones RL (May 1998). "Barley aleurone cells contain two types of vacuoles. Characterization Of lytic organelles by use of fluorescent probes". The Institute Cell. 10 (v): 685–98. doi:10.2307/3870657. JSTOR 3870657. PMC144374. PMID 9596630.
- ^ Holtzman E (1989). Lysosomes. New York: Plenum Printing. pp. 7, 15. ISBN978-0306-four-3126-5.
- ^ De DN (2000). Plant Cell Vacuoles: An Introduction. Australia: Csiro Publishing. ISBN978-0-643-09944-9.
External links [edit]
| | Look upward lysosome in Wiktionary, the free dictionary. |
-
 This article incorporates public domain textile from the NCBI document: "Science Primer".
This article incorporates public domain textile from the NCBI document: "Science Primer". - 3D structures of proteins associated with lysosome membrane
- Hide and Seek Foundation For Lysosomal Research
- Lysosomal Affliction Network, a research consortium funded by the NIH through its NCATS/Rare Diseases Clinical Research Network
- Self-Destructive Behavior in Cells May Agree Primal to a Longer Life
- Mutations in the Lysosomal Enzyme–Targeting Pathway and Persistent Stuttering
- Animation showing how lysosomes are fabricated, and their role
Source: https://en.wikipedia.org/wiki/Lysosome
Posted by: coxyourock.blogspot.com

0 Response to "What Is The Main Function Of The Lysosomes In An Animal Cell?"
Post a Comment